This ‘super antibody’ for COVID fights off multiple coronaviruses
An newly identified immune molecule raises hopes for a vaccine against a range of viruses related to SARS-CoV-2.
You have full access to this article via your institution.
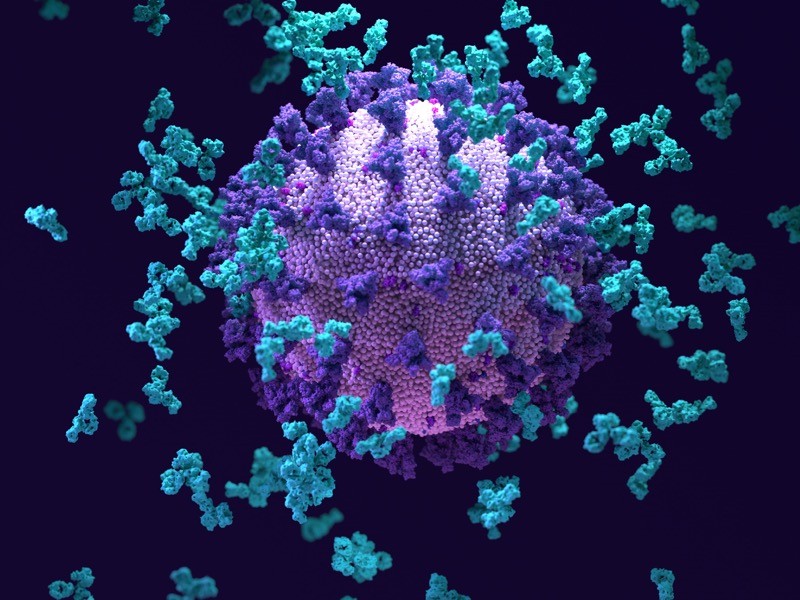
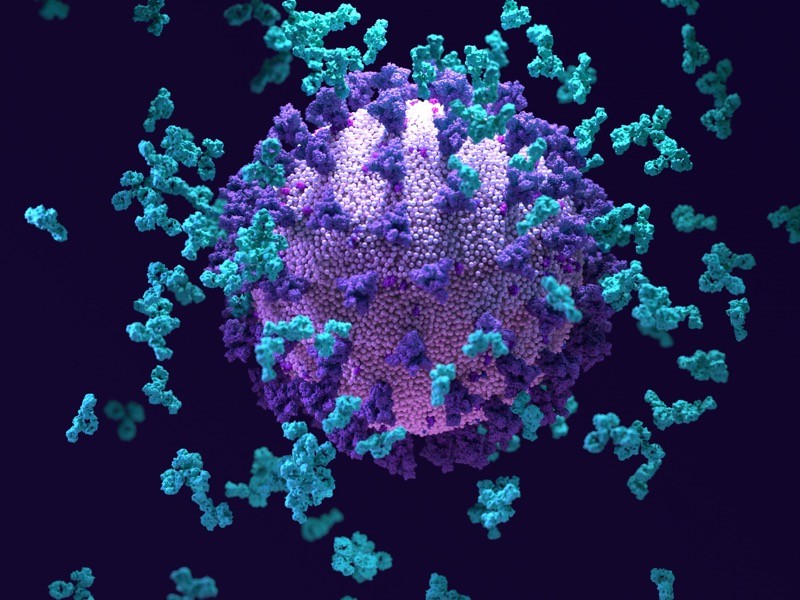
Antibodies (light blue; artist’s impression) swarm around a SARS-CoV-2 particle.Credit: Design Cells/SPL
Scientists have uncovered an antibody that can fight off not only a wide range of SARS-CoV-2 variants, but also closely related coronaviruses
1. The discovery could aid the quest to develop broad-ranging treatments and vaccines.
Tyler Starr, a biochemist at the Fred Hutchinson Cancer Research Center in Seattle, Washington, and his co-authors set out to shed light on a problem facing antibody treatments for COVID-19: some variants of SARS-CoV-2 have acquired mutations that
enable the virus to escape the antibodies’ grasp.
The researchers examined 12 antibodies that Vir Biotechnology, a company based in San Francisco, California, that was involved in the study, isolated from people who had been infected with either SARS-CoV-2 or its close relative SARS-CoV. Those antibodies latch on to a fragment of viral protein that binds to receptors on human cells. Many antibody therapies for SARS-CoV-2 infection grab the same protein fragment, called the receptor binding domain.
The researchers compiled a list of thousands of mutations in the binding domains of multiple SARS-CoV-2 variants. They also catalogued mutations in the binding domain on dozens of SARS-CoV-2-like coronaviruses that belong to a group called the sarbecoviruses. Finally, they assessed how all these mutations affect the 12 antibodies’ ability to stick to the binding domain.
COVID and the brain: researchers zero in on how damage occurs
One antibody, S2H97, stood out for its capacity to adhere to the binding domains of all the sarbecoviruses that the researchers tested. S2H97, which the authors dub a pan-sarbecovirus antibody, was able to prevent a range of SARS-CoV-2 variants and other sarbecoviruses from spreading among cells growing in the laboratory. It was also powerful enough to protect hamsters against SARS-CoV-2 infection. “That’s the coolest antibody that we described,” Starr says.
A closer examination of S2H97’s molecular structure revealed that it targets a previously unseen and well-hidden region on the binding domain — a section that is revealed only when the domain pops up to bind to a cell’s receptor. Starr notes that molecules targeting this binding-domain region could generate protection against multiple viruses, and might one day be used in pan-sarbecovirus vaccines.
The other 11 antibodies could target a variety of viruses, but the more effectively an antibody blocked the entry of the earliest known SARS-CoV-2 strain into a cell, the smaller the range of viruses it could bind. The team also found that antibodies that could disable a wide variety of viruses targeted sections of the binding domain that tended not to change as the virus evolved.
It’s good news that the team has identified antibodies that can bind to a range of sarbecoviruses, says Arinjay Banerjee, a virologist at the University of Saskatchewan in Saskatoon, Canada. “The biggest question that remains is, what about viruses that we don’t know exist yet?”
Although scientists can’t test an antibody’s activity against an unknown virus, Banerjee adds, pan-sarbecovirus treatments and vaccines would help to prepare the world to fight the next coronavirus that jumps from wildlife into humans.
doi: https://doi.org/10.1038/d41586-021-01917-9
UPDATES & CORRECTIONS
- Correction 15 July 2021: An earlier version of this article stated incorrectly that all of the antibodies in the research were from people who had recovered from COVID-19.
References